How cells receive signals from their environment and how they interpret this information, whether to mount a response or not, is a fundamental problem in biology. Frequently, during development, cells are exposed to different levels of signals, or morphogens. In some cases, cells will respond only when signals are above a certain threshold level. We are studying one such cellular response, where cells respond above a certain threshold level by activation of a specific genetic program.
We study this problem during the very first step of neural induction in the ectoderm of ascidian embryos. This takes place at the 32-cell stage of development. Eight anterior ectoderm cells are all exposed to FGF-ligands via cellular contact between ectoderm cells and FGF-expressing mesendoderm cells. FGF signals act via the ERK signalling pathway to activate Otx in the neural precursor (pink cell in Figure. 1).
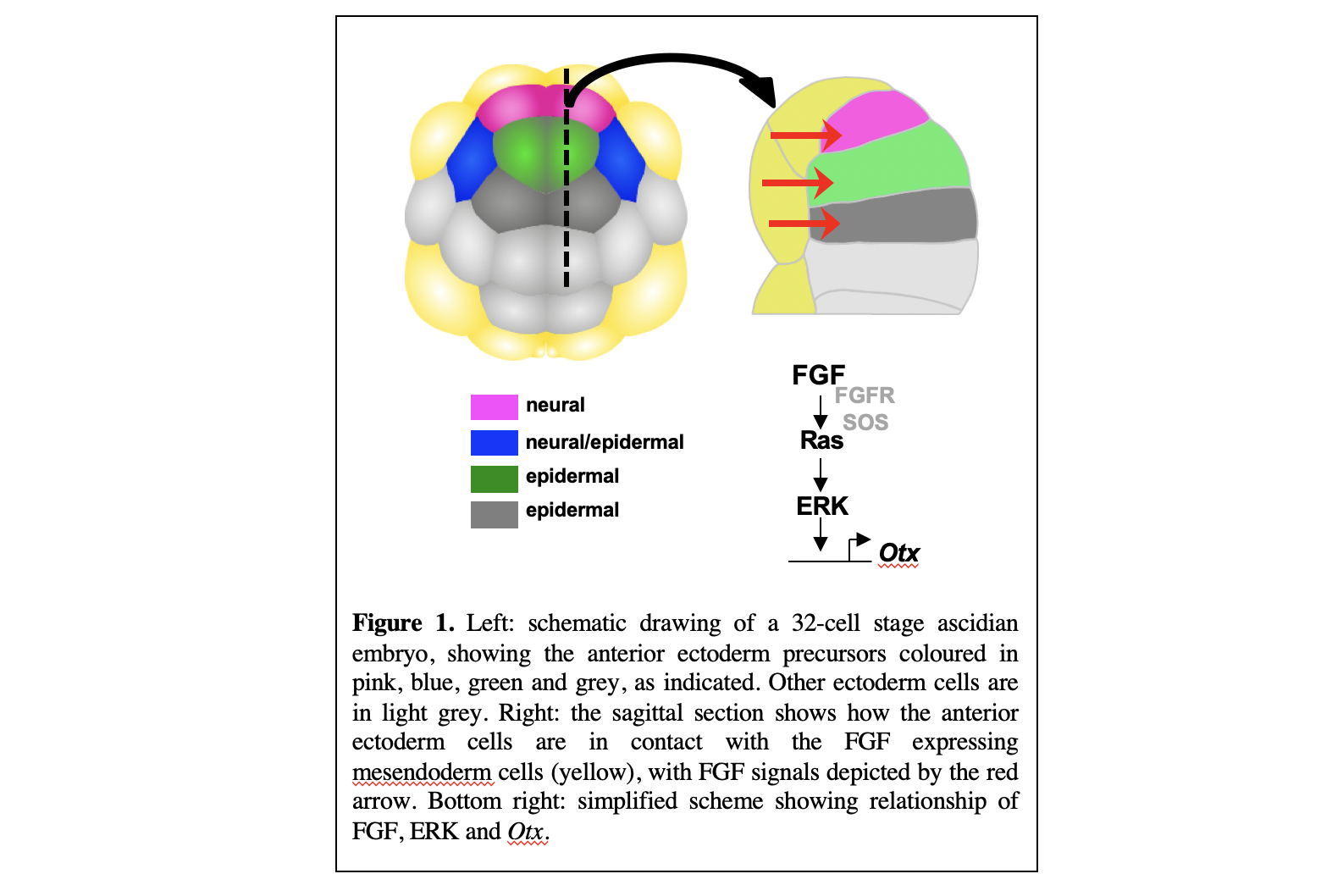
It has been shown that each ectoderm cell has a different area of cell surface contact with the FGF-expressing mesendoderm cells, such that the neural precursors (pink cells in Figure.1) have the largest area of cell contact. We found that these variable levels of FGF signalling input were mirrored in the levels of ERK activity measured in the different cells such that a rather smooth relationship could be measured between the area of the cell in contact with FGF-expressing cells and the levels of ERK activity (Figure 2). ERK activation is then converted into a very steep, quasi-all-or-none, threshold response of Otx transcriptional activation (Figure 2).
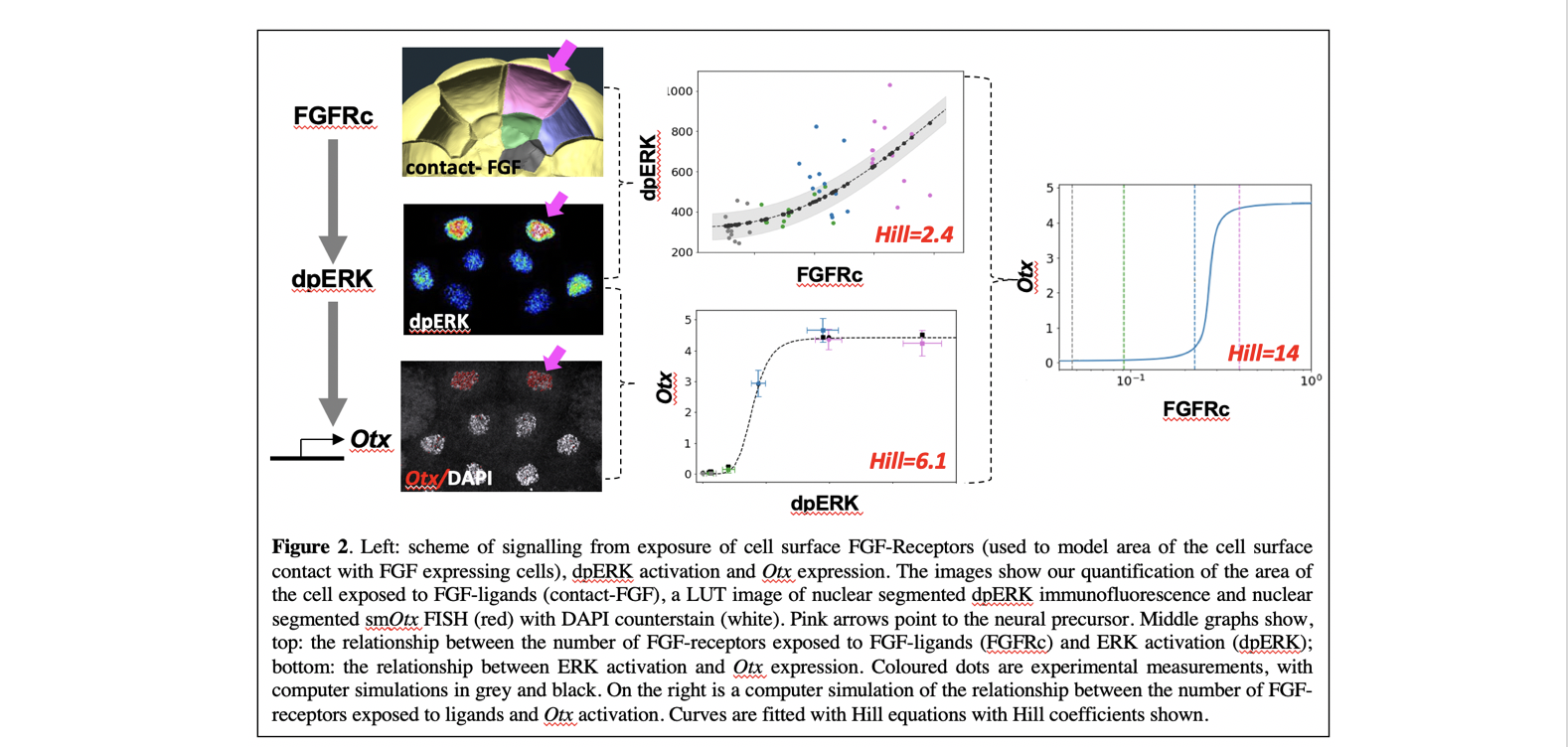
Using our experimental measurements, we constructed a computational model of ascidian neural induction, modelling the steps between FGF and ERK and between ERK and Otx. This helped us to understand how robust the process was to parameter changes and also to measure relationships that were not possible to measure experimentally, such as the relationship between the cell exposure to FGF ligands and the transcriptional response of the Otx gene (Figure 2, right).
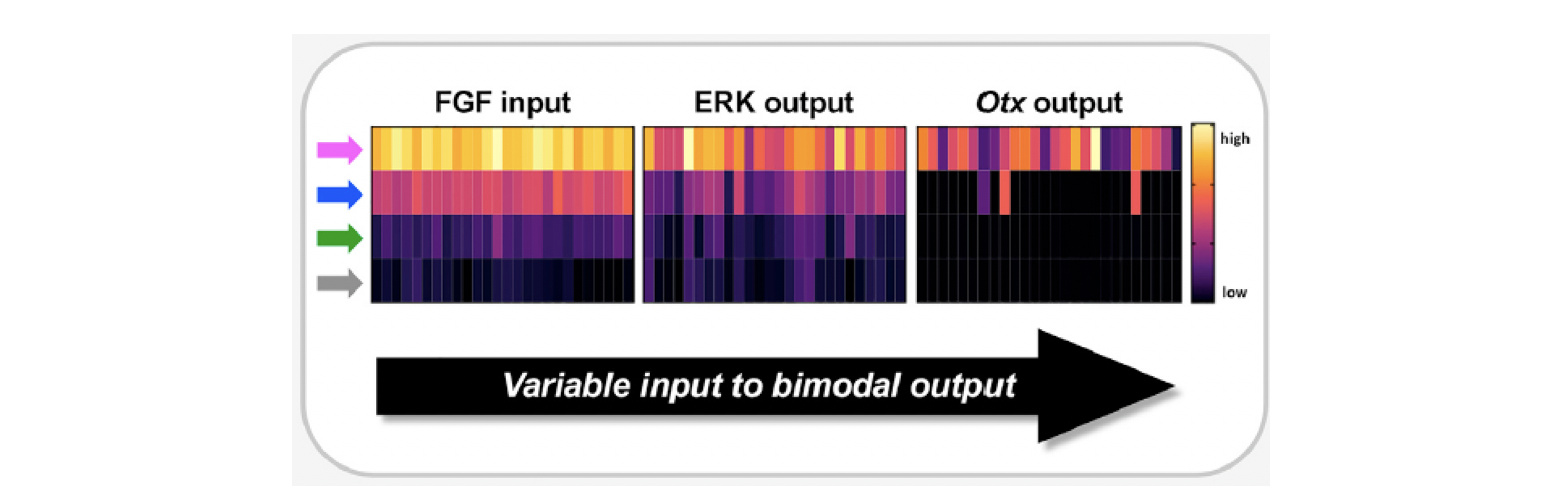
In summary, our results showed that much of the all-or-none response during early neural induction, takes place not between FGF-contact and ERK activation, but between ERK activation and Otx expression (Figure 2). During this process therefore, graded signalling inputs are converted to more digital outputs (Figure 3).
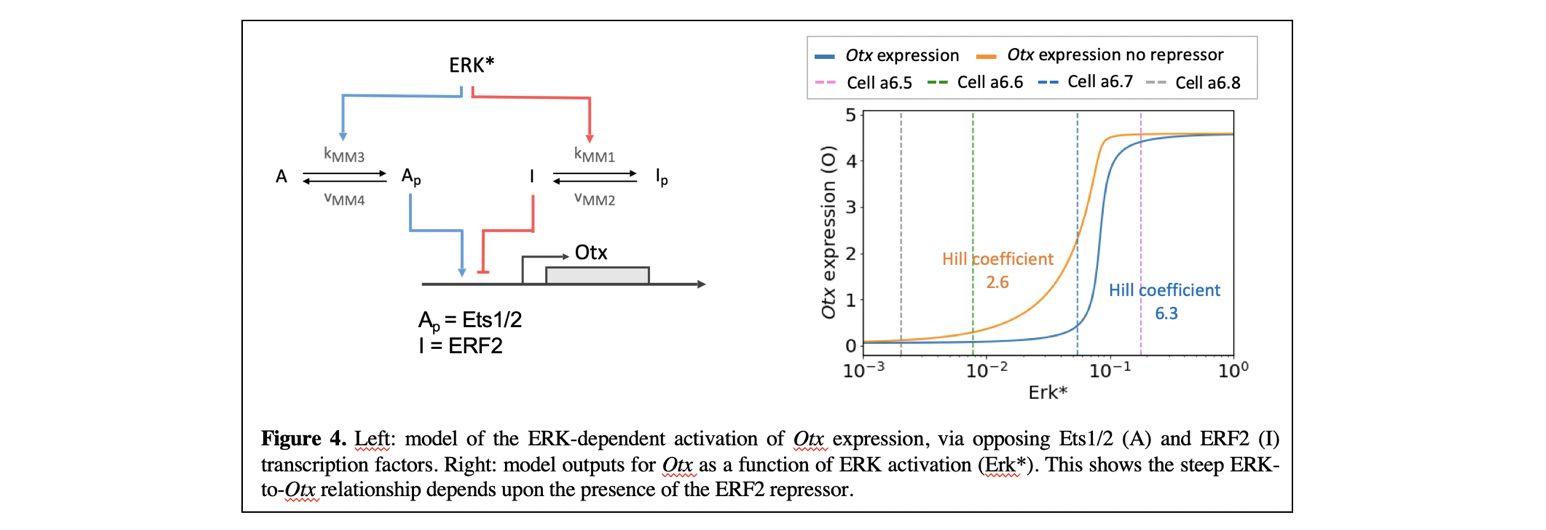
We are now trying to understand in detail how the steeply sigmoidal transcriptional response is made. Otx is induced as a direct response to FGF signals, which activates transcription via the ETS transcriptional activator Ets1/2, which binds to ETS sites in the upstream regulatory sequences of Otx (Bertrand et al., 2003). We have shown that the correct Otx expression output also depends upon the presence of a repressor of the ETS family of transcription factors, ERF2. ERF factors bind to the same binding sites as Ets1/2, but are negatively regulated by ERK via nuclear export (Le Gallic et al., 1999; Le Gallic et al., 2004). Thus, downstream of ERK are two transcription factors of the ETS family, an activator Ets1/2 and a repressor ERF2, which are inversely regulated by ERK activation and that are predicted to compete for the same binding sites in the Otx upstream sequences (Figure 4). Our experiments and computational models shows that ERF2 is critical for the steepness of the relationship between ERK and Otx (Figure 4) (Bettoni et al., 2023; Williaume et al., 2021).
This project is a collaboration between our group and Geneviève Dupont (Faculty of Science, Université Libre de Bruxelles), Sophie de Buyl (Department of Physics of the Vrije Universiteit Brussel), Yutaka Satou (Kyoto University), Kaoru Imai (Osaka university).
Publications from this study:

Laboratoire de Biologie du développement UMR 7009
Cette UE se déroule sur 2 semaines et a lieu au laboratoire de Biologie du Développement de Villefranche -sur-Mer (LBDV). Elle inclut l’examen de l’UE d’analyse scientifique (5V089) suivie par les étudiants de la spécialité de Biologie du Développement.
Durant la 1ère semaine, les étudiants participent à des ateliers et des rencontres avec les chercheurs du laboratoire.
Durant la 2ème semaine, les étudiants sont répartis dans les équipes pour y réaliser un mini projet qu’ils présentent le dernier jour du cours. Le cours est donné en anglais pour tout ou partie.
Modalités d’évaluation
Présentation orale du mini-projet (binôme, 100 %)
UE en anglais (partiellement ou totalement)
Master BMC, S3, 6 ECTS
Code UE: 5V200
Responsable de l’UE: Carine BARREAU (MCU): carine.barreau [at] obs-vlfr.fr
Cette UE permet aux étudiants de 1ère année de Master de passer 2 semaines à l’Observatoire Océanologique de Villefranche-sur-Mer. Le cours est obligatoire pour les étudiants du Master Biologie Intégrative, parcours Biologie et Bioressources Marines (BBMA) tandis qu’il peut être choisi en option par les étudiants du Master Biologie Moléculaire et Cellulaire (BMC). Les étudiants participent à des ateliers de présentation des organismes marins utilisés par les équipes de recherche du laboratoire (LBDV) et apprennent à les manipuler au cours de travaux pratiques dont les thématiques vont de la Biologie du Développement fondamentale à la toxicologie appliquée.
Modalités d’évaluation
Analyse d’article et présentation orale (50%)
Compte-rendu écrit de TP (50%)
UE partiellement en anglais
Master BIP, S2, 6 ECTS
Code UE: 4B022 (ouverte au Master BMC)
Responsable de l’UE: Carine BARREAU (MCU): carine.barreau [at] obs-vlfr.fr
Cette UE complémentaire se déroule sur 2 semaines et permet aux étudiants de découvrir les différents aspects (métiers & recherche) de l’Observatoire Océanologique de Villefranche-sur-Mer (OOV). Ateliers et journal clubs sont organisés afin que les étudiants mettent en pratique leurs connaissances théoriques en biologie et développent leur capacité de communication scientifique en français et en anglais.